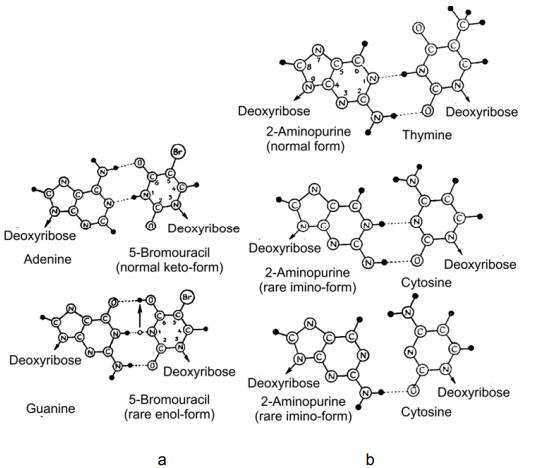
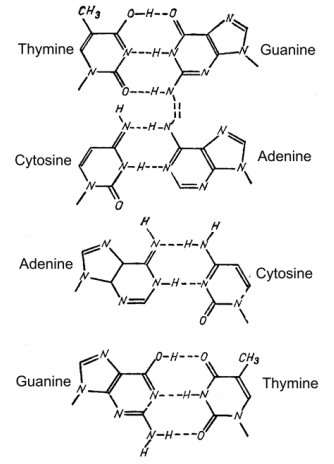
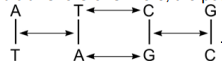
Page 162
Reparation Processes
The primary damages and changes occurred in the genetic material are
only a stage of the mutation process. Not less important is the next stage
connected with the further “fate” through the “laboratory labyrinths” of cell
structures determining the heredity. Practice and experiments show that not
all of them are firmly included in the genetic material and complete with
mutations. Obviously, there exist mechanisms which repare or remove
them in some way, thus maintaining the normal status (status quo) of
organisms. These processes have been called reparation.
If bacterial cells are subjected to the action of high doses of UV-radiation
(~280 nm), their ability to form colonies abruptly decreases. It has been
observed that in some bacteria this ability is restored after their exposure to
daylight, that has led to the discovery of phenomenon photoreparation or
photoreactivation. For the first time it is observed by Kelner (1949 a, b) at
lighting up Actinomyces suspension, but it is confirmed on a number of other
microorganisms — bacteria, phages, paramecia, etc.
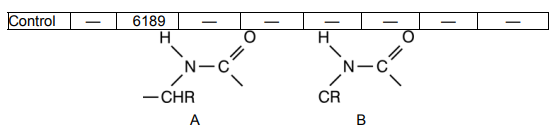
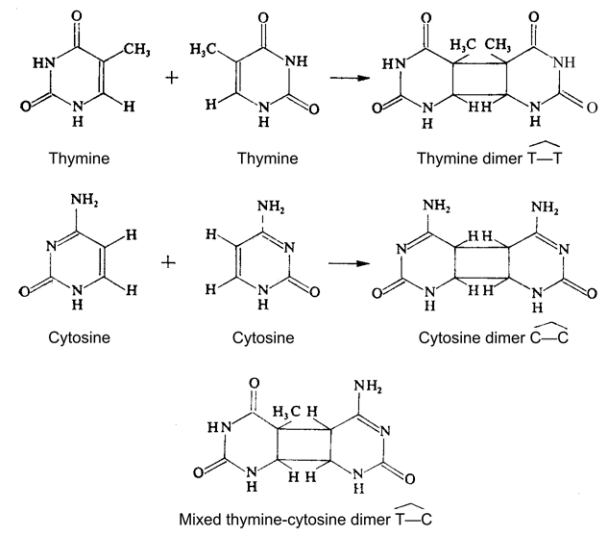
It has been established that by UV-irradiating dimers of thymine (see
Fig. 2–80), are formed in DNA, which disturbs the structure and functions of
the genes. Except between thymine bases (TT), dimers may occur between
uracil and cytosine (UC) and only between cytosine (CC). The more are the
dimers formed, the greater is the lethal effect. After photoreactivation the
dimers disappear (Setlow. Setlow. 1962).
The beginning of more profound studies on the processes of reparation
is laid by revealing the enzyme photoreactivation (Setlow, Setlow, 1963) and
elucidating the mechanism of so-called dark reparation of the damages in
DNA. Many bacteria repair the damages caused by UV-rays in dark. That has
led to the supposition of existence of different reparation mechanisms. Some
bacteria have shown greater susceptibility to radiation, other have been
found more resistant. It proved that during dark reparation without any
assistance the resistant lines remove pyrimidine dimers in DNA, while the
susceptible ones do not remove them (Setlow, Carrier, 1964; Boyce, Howard
Flanders, 1964; Howard-Flanders, 1968. 1973). According to Auerbach
(1976) photoreparation can achieve 100% effectiveness and correctness of
the transformation of dimers into monomers.
On the basis of performed investigations, the mechanisms of
reparation are reduced to three types: a) photoreactivation; b) reparation
through cutting (excision reparation); c) postreplication reparation.
Photoreactivation is the most simple mechanism, since only one
enzyme is required that should be capable to “recognize” and bind the part
of DNA (thymidine dimer) underwent a primary damage. The source of
energy is visible light, which serves the photoreactivating enzyme in
separating the dimer and restoring the initial state. It is established that
daylight is most effective between 310 and 440 nm (Setlow. 1966).